About
I am a Chicago DFI Fellow and postdoctoral associate at the University of Chicago. I completed my Ph.D. in Bioinformatics and Computational Biology at the University of Minnesota - Twin Cities.
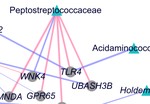
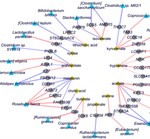
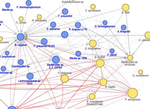
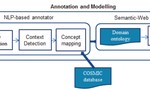
My research focuses on using machine learning approaches for understanding host-microbiome interactions. I am developing machine learning tools to integrate multi-omics and clinical data to shed light on the intricate links between the microbiome, clinical variables, and disease. As part of my doctoral research, I developed and applied a machine learning-based framework to perform a comprehensive characterization of interactions between the gut microbiome and host gene expression in patients with colorectal cancer, inflammatory bowel disease, and irritable bowel syndrome.
Interests
- Bioinformatics
- Host-microbiome interaction
- Machine Learning
- Multi-omics
Education
-
PhD in Bioinformatics and Computational Biology, 2021
University of Minnesota
-
MS in Computer Science, 2016
Lehigh University